From 2952dc03f5427bdaa26fb41a861d6a0ca052fe47 Mon Sep 17 00:00:00 2001
From: Calvin <179209347@qq.com>
Date: Sun, 13 Feb 2022 19:08:17 +0800
Subject: [PATCH] upgrade milvus to v2.0
---
6_biomedicine/dna_sequence_search/README.md | 79 +--
.../dna-search-ui/package.json | 2 +-
.../dna_sequence_search.iml | 319 ----------
.../dna_sequence_search/pom.xml | 2 +-
.../me/aias/common/milvus/ConnectionPool.java | 235 ++++++++
.../aias/common/milvus/MilvusConnector.java | 37 --
.../me/aias/config/MilvusConfiguration.java | 26 -
.../me/aias/config/ModelConfiguration.java | 2 +-
.../me/aias/controller/DNASeqController.java | 41 +-
.../me/aias/controller/SearchController.java | 51 +-
.../java/me/aias/service/SearchService.java | 103 ++--
.../aias/service/impl/SearchServiceImpl.java | 560 +++++++++++-------
.../main/java/me/aias/tools/MilvusInit.java | 107 +++-
.../src/main/resources/application.yml | 28 +-
.../molecular-search/molecular-search.iml | 207 -------
15 files changed, 773 insertions(+), 1026 deletions(-)
delete mode 100644 6_biomedicine/dna_sequence_search/dna_sequence_search/dna_sequence_search.iml
create mode 100644 6_biomedicine/dna_sequence_search/dna_sequence_search/src/main/java/me/aias/common/milvus/ConnectionPool.java
delete mode 100644 6_biomedicine/dna_sequence_search/dna_sequence_search/src/main/java/me/aias/common/milvus/MilvusConnector.java
delete mode 100644 6_biomedicine/dna_sequence_search/dna_sequence_search/src/main/java/me/aias/config/MilvusConfiguration.java
delete mode 100644 6_biomedicine/molecular_search/molecular-search/molecular-search.iml
diff --git a/6_biomedicine/dna_sequence_search/README.md b/6_biomedicine/dna_sequence_search/README.md
index 8485e625..55ec1f34 100644
--- a/6_biomedicine/dna_sequence_search/README.md
+++ b/6_biomedicine/dna_sequence_search/README.md
@@ -5,7 +5,7 @@ http://aias.top/
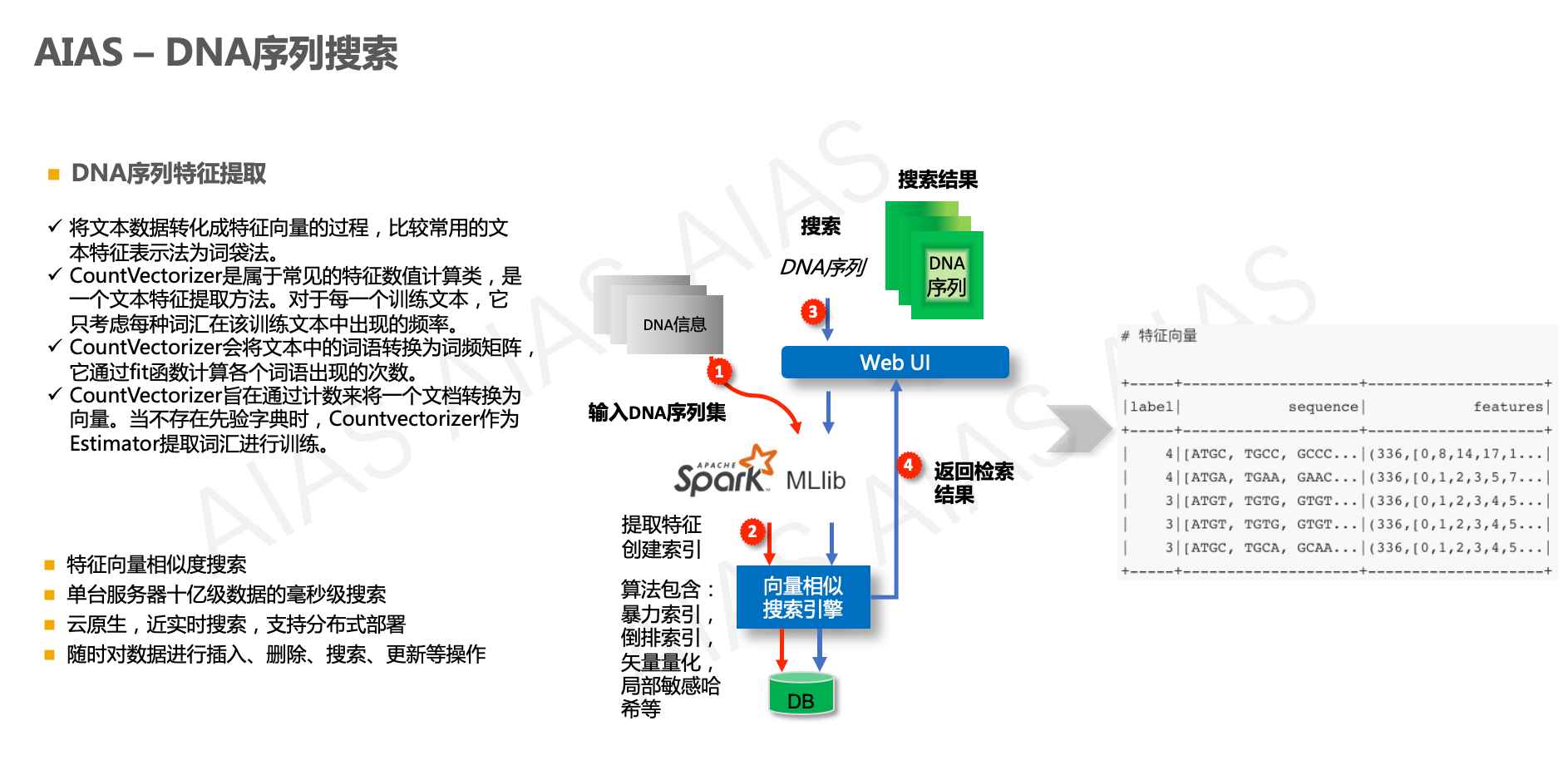
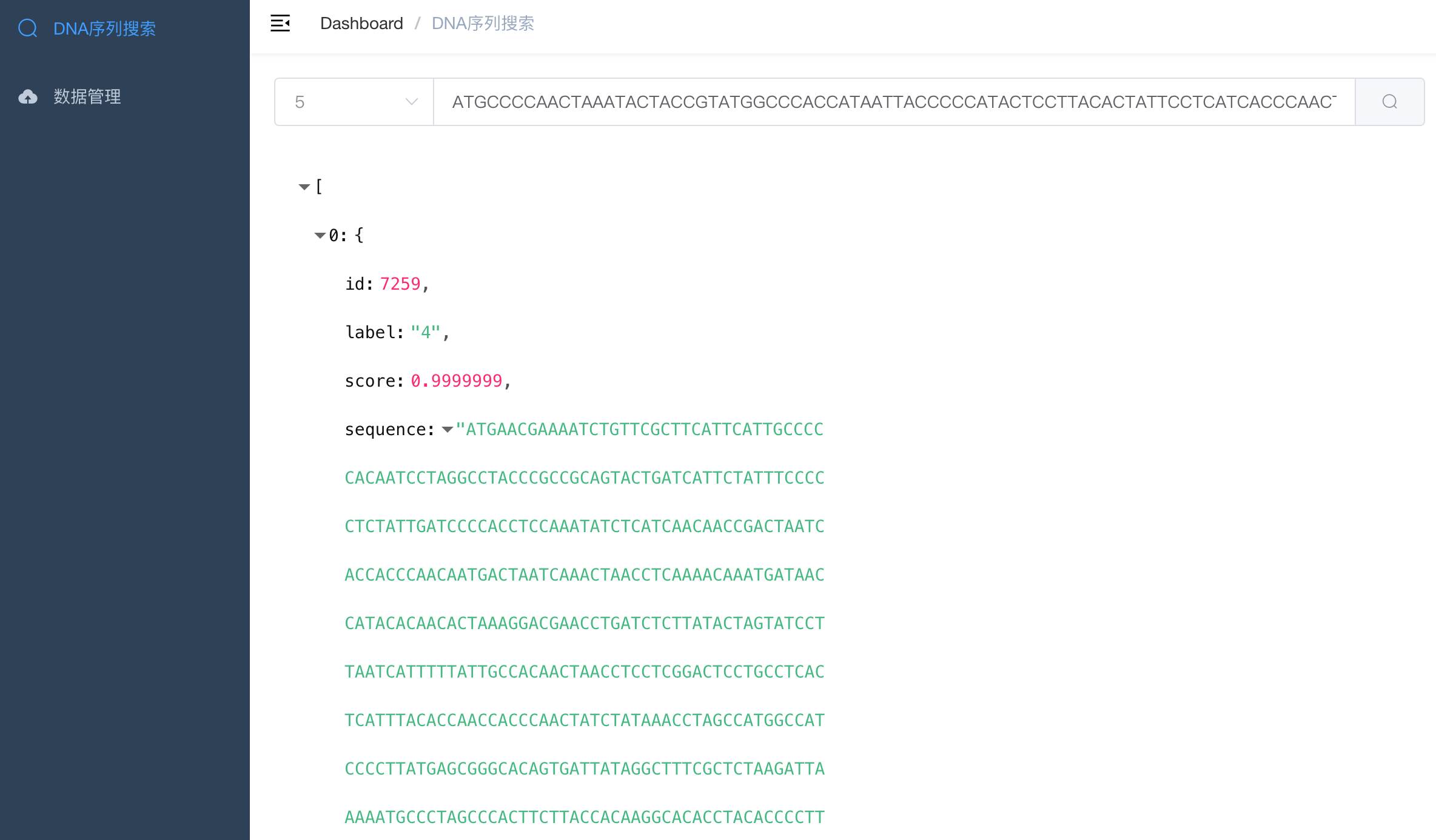
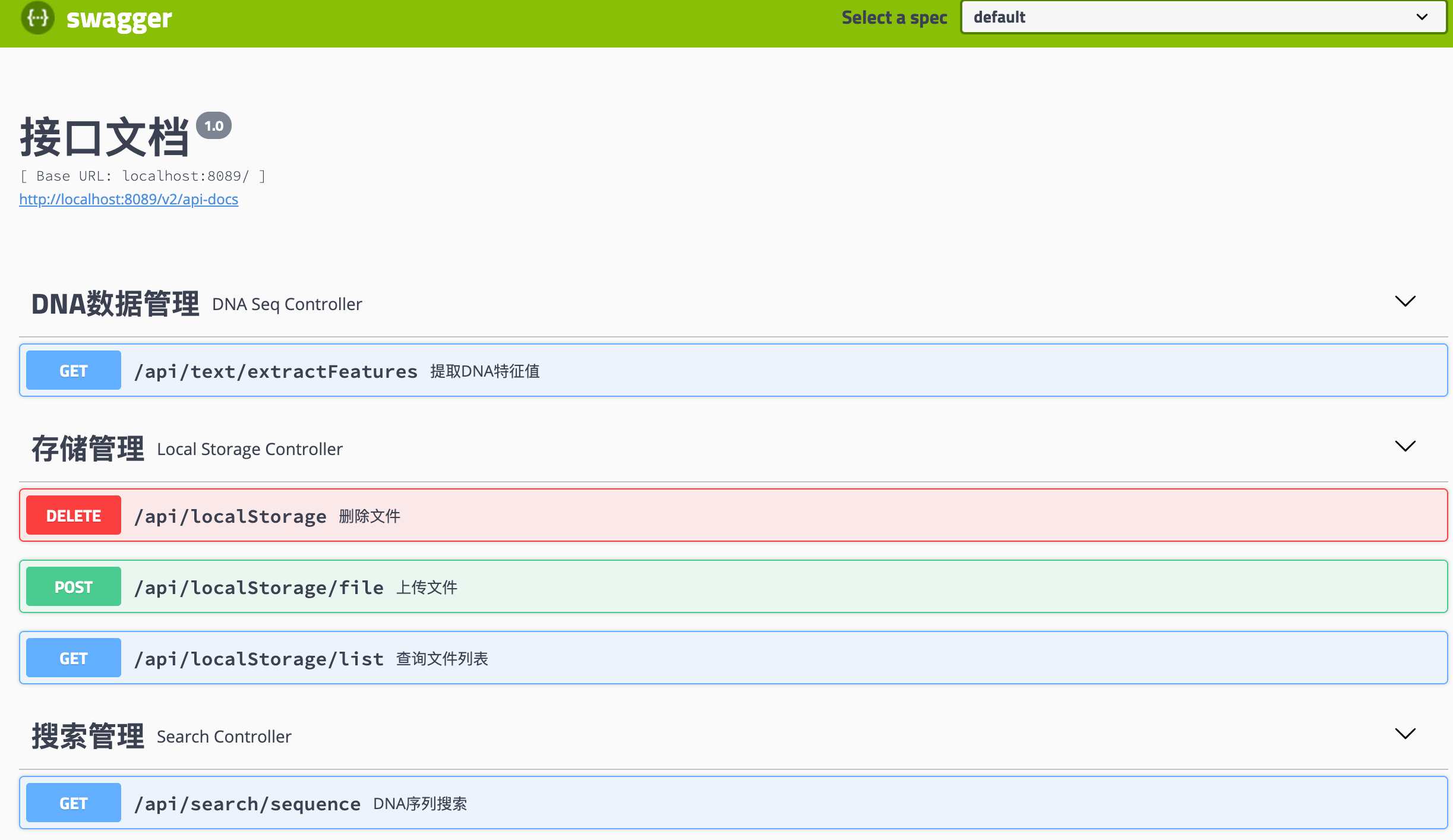
本例子提供了DNA序列搜索,支持上传文件文件,使用spark mlib计算模型提取特征,并基于milvus向量引擎进行后续检索。
-

+
-

+

-

+

-

+

-

+
-

+
 +
+
 +
+
 +
+
 +
+
 +
+
 +
+
 +
+